Home
Optimizing Pandas
Introducing vectorization
 Pandas!! By Chi King, CC BY 2.0
Pandas!! By Chi King, CC BY 2.0
Here's a quick trick to speed up processing time, when performing
analysis on a Pandas DataFrame. Say we
want the sum of all the values in specific rows or columns. Even for such a simple
task, there are
several ways to approach it. So what are our options?
Let's consider a few: one horrible, and a few decent. And to conclude, we'll find there's an even more computationally efficient way to perform operations over an entire dataframe; we'll see how to easily access NumPy's optimized C source code. But first, let's consider our worst possible option.
Note: we're using some Jupyter cell
magic, %%time to time our function calls. This
function measures runtime on a given cell. These times vary, but in general,
we can use these times as a baseline measurement for performance. Time is
measured in seconds, milliseconds, or microseconds.
Option 1: The Dreaded
for loop
The naive approach is to loop over the entire
DataFrame, using for or
while. Let's see how this performs.

This a relatively small dataset , so 3 seconds to perform this function could mean a much slower down on larger sets. This approach doesn't utilize panda's built in methods; these tend to have more optimized performance. Even better, I find these methods easier to conceptualize when coding. This gained efficiency will critical when working with massive datasets. Let's consider better options.
Option
2:df.apply()
Now for our next best solution. Let's get
familiar with df.apply(). The beauty of
apply() is its flexibility. You can pass any NumPy
operation, such as np.sqrt(), to perform
calculations over the DataFrame.
Even more impressive, you can write custom functions and pass them to
apply() for context specific operations, such as
calculating conversion rates or other Key Performance Indicators (KPIs).
Let's see how to use it for our simple task. Here we sum over
rows.
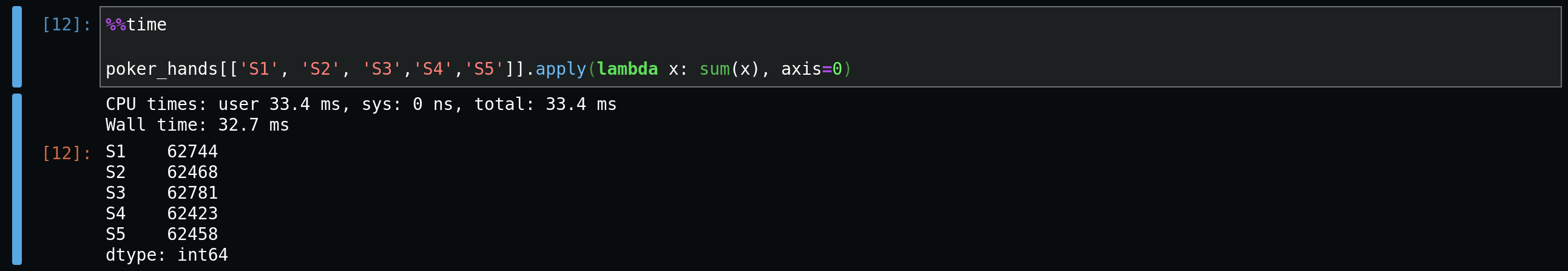
To specify whether the calculation is performed over columns or rows, you
pass the argument axis=0, for rows, and
axis=1, for columns.
Option
3:df.sum()
Now, we'll start using a more optimized approach,
df.sum(). Here we get a better runtime. The cost
however, is we don't get the flexibility of
apply() to pass custom or NP functions.
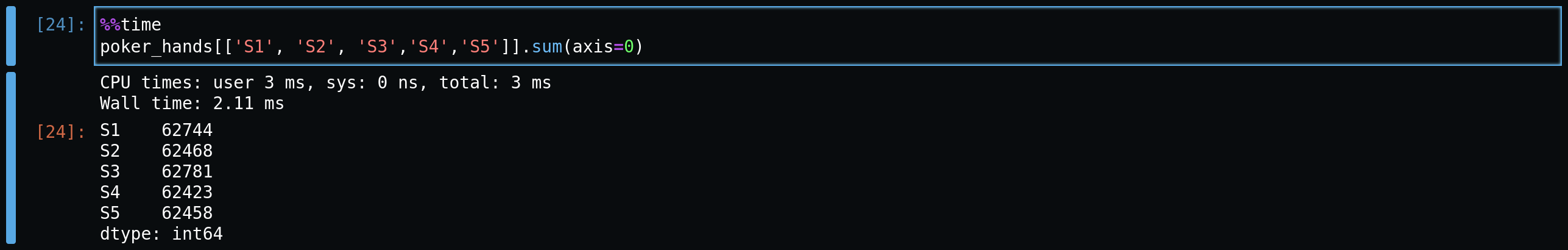
Again we specify the axis to sum over rows. Incredibly, we see a huge jump
in performance by using sum. But can we go
further?
Option 4: Vectorizing NumPy Arrays using
df.values
Now for the moment we've all been waiting for, our champion,
.values
NumPy, under the hood, uses optimized C code. When we call a NumPy function, we're actually accessing the Python binding of a C library. This neat feature is what has really enabled state of the art data analysis and data science in Python. Python, the language, was consciously designed to favor code that's more beautiful for the developer than the computer. Due to the C bindings, we get the best of both code 'readability' and performance! Specifically, the C library is performing a powerful method of computation over large numbers of arrays, called vectorization.
But notice, we're talking about NumPy arrays here, not Pandas
DataFrames. Pandas uses a native Python coded
series object for its operations; NumPy uses
ndarray. These ndarray
objects leave out a lot of 'nice to haves' found in
pd.series. These include data-type checking, indexing,
and other potentially expensive features. Without these,
ndarrays are many times faster and can be quite
useful, when the additional functionality is not necessary.
Let's see how to work with a DataFrame as an
ndarray.

It's as simple as specifying df.values, which
is an attribute of the DataFrame. Well,
technically it's a property, which you can think of as an attribute you
can't (or shouldn't) modify externally as a developer. You specify
.values and then and call a method over it.
Alternatively, you can call df.to_numpy for
similar functionality. And a word of caution, if your column or row contains
a mix of both ints and
floats, one type will be determined to capture all of
the types. Floats being a lot more memory
intensive could lead to a possible issue you didn't foresee, so plan
your dtypes accordingly.
Concluding Remarks
And there we have it! Compared to where we started with the
for loop and even apply,
we've seen dramatic improvements, of orders of magnitude. When running
these operations on massive datasets, we can expect even larger differences
in performance.
I hope this article helped you see how Pandas offers the blessing of a variety of ways to solve a task. With a little insight into how Pandas functions under the hood, you can find the approach that cuts down on precious computation time and resources. Now go on and write smarter and faster Python code!
Written on September 2 by Feli Gentle.